Receptor Summary
10 20 30 40 50 60 70 80 90 100
MFSPADNIFI ILITGEFILG ILGNGYIALV NWIDWIKKKK ISTVDYILTN LVIARICLIS VMVVNGIVIV LNPDVYTKNK QQIVIFTFWT FANYLNMWIT
110 120 130 140 150 160 170 180 190 200
TCLNVFYFLK IASSSHPLFL WLKWKIDMVV HWILLGCFAI SLLVSLIAAI VLSCDYRFHA IAKHKRNITE MFHVSKIPYF EPLTLFNLFA IVPFIVSLIS
210 220 230 240 250 260 270 280 290 300
FFLLVRSLWR HTKQIKLYAT GSRDPSTEVH VRAIKTMTSF IFFFFLYYIS SILMTFSYLM TKYKLAVEFG EIAAILYPLG HSLILIVLNN KLRQTFVRML
310 320 330 340 350 360 370 380 390 400
TCRKIACMI
Protein Diagram
When using this 2D protein diagram,please cite:
Protter: interactive protein feature visualization and integration with experimental proteomic data.
Omasits U, Ahrens CH, Müller S, Wollscheid B.
Bioinformatics. 2014 Mar 15;30(6):884-6. doi: 10.1093/bioinformatics/btt607
Click here to view the paper:
Protter paper
Citations:
Promiscuity and selectivity of bitter molecules and their receptors. Bioorg Med Chem 23 (2015) 4082-4091.
doi: 10.1016/j.bmc.2015.04.025
3D Visualizer: NGL, A WebGL Protein Viewer
AS Rose and PW Hildebrand. NGL Viewer: a web application for molecular visualization. Nucl Acids Res (1 July 2015) 43 (W1): W576-W579.
doi: 10.1093/nar/gkv402
Jumper, J., Evans, R., Pritzel, A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). doi: 10.1038/s41586-021-03819-2
Mihaly V., Stephen A., Mandar D. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models, Nucleic Acids Research, Volume 50, Issue D1, 7 January 2022, Pages D439–D444, doi: 10.1093/nar/gkab1061
Category
Name
PUBCHEM
Structure
Reference
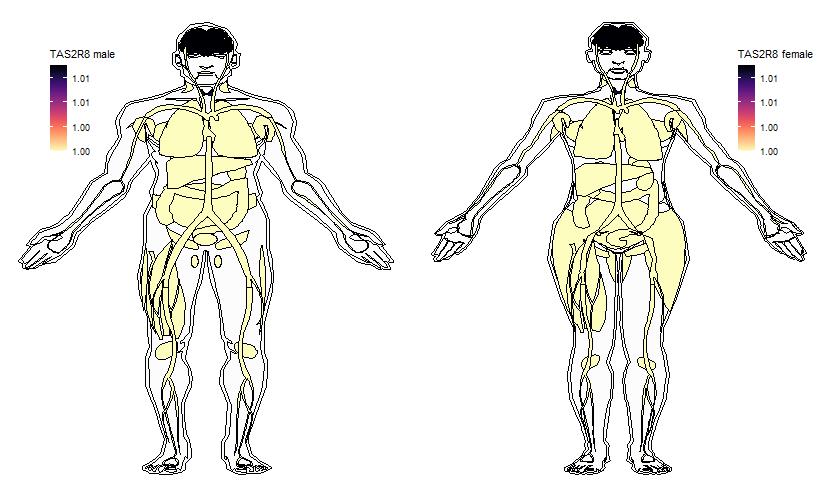
Ectopic expression of some Receptors in a variety of tissues was drawn using R gganatogram package, based on a comprehensive expression analysis with Next Generation Sequencing (NGS) technique (View more). Expression data are taken from the table-Expression profile of all genes in 16 different human tissues. Original values are added by one to show a log10 transformed profile.